June 8-11, 2019
The 17th Annual Meeting of the Complex Trait Community, in collaboration with the Rat Genomics Community, will be held in La Jolla, California on the University of California San Diego campus from Saturday, June 8 to Tuesday, June 11, 2019.
The only activity on June 8 will be an informal welcome reception in the evening (see below).
The meeting will cover quantitative approaches to mouse and rodent genetics (other model organisms also welcome). We will have three plenary speakers, poster session, and many short talks that will be drawn from submitted abstracts and organized thematically.
We will have a closing dinner on Tuesday, June 11, at the Kellogg Park at the beach (map).
There will be no registration fee, thanks for the support of our sponsors, GENCOVE, iGenomX, and 10xGenomics.
Abstract submission is closed.
If you submitted an abstract but did not receive an email indicating that your abstract had been selected (for a talk or a poster presentation), please email us.
If you can no longer attend the meeting, please let us know by email at your earliest convenience.
Plenary session speakers:
Sunday, June 9, 8:30 am: Trey Ideker, PhD, Professor of Medicine, Adjunct Professor of Bioengineering & Computer Science, University of California San Diego.
Monday, June 10, 8:30 am: Bing Ren, PhD, Professor of Cellular and Molecular Medicine, University of California San Diego.
Tuesday, June 11, 8:30 am: Melissa Gymrek, PhD, Assistant Professor, Department of Medicine, University of California San Diego.
Poster size:
Each presenter will have poster board space 48″ by 48″ wide. Poster of any standard size will fit.
Program:
Saturday June 8
6pm – 8 pm Welcome reception (drinks and appetizers)
Location: Biomedical research Facility II (BRFII), Room 1104 (first floor)
Sunday June 9
Location: Sanford Medical Education and Telemedicine Center, Auditorium (first floor)
8.30 – 9.30 Invited Talk: Trey Ideker “Decoding complex genetics with protein network models of the cell”
9.30 – 10.30 Short talks (12 min+3 min QA)
Session chair: Clarissa Parker
Abraham Palmer “First five years of the NIDA Center for GWAS in Outbred Rats”
Sarah Heuer “Multi-omic analysis identifies transcriptional networks and drivers associated with cognitive aging and Alzheimer’s disease”
Thom Saunders “Generation and Characterization of Transgenic Rats Carrying Cre Recombinase Knockins at the Dopamine D1 and Adenosine 2a Receptor Loci”
Vivek Kumar “Quantitative genetics of ethological endophenotypes in the laboratory mouse”
10.30 – 11 Coffee break
11 – 12.30 Short talks (12 min+3 min QA)
Session chair: Elissa Chesler
Aron Geurts “Hybrid Rat Diversity Program”
Lauren Vanderlinden “Examining the influence of population structure and genetic diversity within the Hybrid Rat Diversity Panel on the brain transcriptome and its eQTL”
Shur-Jen Wang “Linking phenotypic values with genomic variations at Rat Genome Database”
Vivek Philip “Mouse Phenome Databse: New tools for curate and integrated primary mouse phenotype data”
Pjotr Prins “Genenetwork/GEMMA and multidimensional data”
Robert Williams “Predictive genetics and the use of massive mouse and rat diallel crosses (DX)”
12.30 – 2 Lunch break (no lunch provided)
2 – 3.30 Short talks (12 min+3 min QA)
Session chair: Richard Mott
John Edgar French “Diet-induced Diversity Outbred mice weight gain followed by calorie restriction weight loss quantitative traits”
Gary Churchill “The Genetic Architecture of Insulin Secretion”
Adriana Huertas-Vazquez “Identification of DNA variants influencing metabolite levels in non-alcoholic fatty liver disease”
Karthickeyan Chella Krishnan “Systems Genetics Approach Identifies Sex-Specific Gene Regulatory Networks in Adipose and Liver Mitochondria”
Leah Solberg Woods “Identification of novel genes for adiposity and lipids using outbred heterogeneous stock rats”
Apurva Chitre “Genome wide association study in 3,173 outbred rats identifies 31 loci for body weight, body mass index, adiposity, and fasting glucose”
3.30 – 4 Coffee break
4 – 5.30 Short talks (12 min+3 min QA)
Session chair: Clement Chow
Brie Katherine Fuqua “A Systems Genetics Approach to Identify Genes and Pathways Involved in Liver Iron Overload and Pathology”
Martin McBride “Extracellular vesicles isolated from osteopontin transfected conditioned media increase H9c2 cell size”
Wei-Zen Wei “Identification of genes associated with time to spontaneous mammary tumor onset and tumor growth rate in BALB NeuT x Diversity Outbred F1 mice”
Jian-Hua Mao “Diverse tumour susceptibility in Collaborative Cross mice: identification of a new mouse model for human gastric tumourigenesis”
Evan Williams “A Multi-Omics Population Approach for Understanding the Aging Mouse Liver”
Isabela Gerdes Gyuricza “Differential gene and protein expression from the aging heart of Diversity Outbred mice”
6 – 8pm Optional workshop on GeneNetwork led by Pjotr Prins (pizza provided by organizers)
Monday June 10
Location: Sanford Medical Education and Telemedicine Center, Auditorium (first floor) for Talks or Rooms 141-145 (first floor) for poster session
8.30 – 9.30 Invited talk: Bing Ren “Genome-wide analysis of Enhancers in Development and Disease”
9.30 – 10.30 Short talks (12 min+3 min QA)
Session chair: Megan Mulligan
Jason Bubier “The relationship between host genetics, microbiome composition and addictive or addictive-predictive behaviour”
Oksana Polesskaya “Gene-microbiome interactions in drug abuse”
Karl Broman “Identification of sample mix-ups and mixtures in microbiome data in Diversity Outbred mice”
Hao Li “Identifying gene function and module connections by the integration of multi-species expression compendia”
10.30 – 11 Coffee break
11 – 12.30 Short talks (12 min+3 min QA)
Session chair: Robert Williams
Hao Chen “Identifying assembly errors in the rat reference genome”
Jun Li “Efforts to create reference quality rat genomes”
David Ashbrook “Sequencing the BXD family, a cohort for experimental systems genetics and precision medicine”
Ryan Lusk “Accuracy of transcriptome reconstruction from simulated RNA sequencing data for identifying exon structure and transcription stop sites”
Harry Smith “Assessing accuracy of de novo transcriptome reconstruction in rat brain by comparing with high quality Iso-Seq complete reads”
Thamar Jessurun Lobo “Evaluation of different transcriptome mapping strategies for the analysis of hybrid mice “
12.30 – 1.30 Lunch break (no lunch provided)
1.30 – 3 Short talks (12 min+3 min QA)
Session chair: Martin McBride
Irit Gat-Viks “Inter-individual diversity of cell-state composition during influenza infection”
Olivia Sabik “Expanding our understanding of the complex genetic architecture underlying osteoporosis using networks”
Basel Al-Barghouthi “Systems Genetics Analysis of Bone Strength in the Diversity Outbred”
Caroline Manet “Host genetics control susceptibility to Zika virus in Collaborative Cross mice”
Stuart Macdonald “Genomewide expression analysis of the adult female gut in the Drosophila Synthetic Population Resource”
Xavier Montagutelli “A loss-of-function mutation in Itgal contributes to the high susceptibility of strain CC042 to Salmonella infections”
3 – 3.30 Poster blitz (1 minute per poster) (auditorium)
Session chair: Laura Saba
List of posters with abstracts
Poster #1: Andrew Deighan “Hematologic Predictors of Lifespan in Diversity Outbred Mice”
Poster #2: Andrew Ouellette “Comparative analysis of working, short and long-term fear memory traits in the BXD and CC genetic reference mouse populations”
Poster #3: Danny Antaki “Mapping Genetic Modifiers of Early Neural Development with the Diversity Outbred Stock”
Poster #4: Douglas Adams “Genome-Wide Association Study of Skeletal Mechanical Integrity using the Heterogeneous Stock Rat”
Poster #5: Duy Pham “Genetic Drivers of Protein Abundance in Pancreatic Islet Cells”
Poster #6: Jennifer Mahaffey “Array and Computational Methods to investigate a role for circRNAs in complex traits in the Hybrid Rat Diversity Panel.”
Poster #7: Jeremy Li “Comparing low-pass sequencing and genotyping for trait mapping in pharmacogenetics”
Poster #8: Jianjun Gao “Adapting genotyping-by-sequencing and variant calling for heterogeneous stock rats”
Poster #9: Keith Brown “RipTide™ High Throughput NGS Library Prep for Genotyping in Populations”
Poster #10: Kian Waddell and Michael Caminear “Genome-wide association for body weight in the diversity outbred mouse population”
Poster #11: Masahide Asano “The fourth term of the National BioResource Project-Rat in Japan and development of rat reproductive technologies”
Poster #12: Matt Dean “The genetic basis of size and shape variation in rat bacula”
Poster #13: Monika Tutaj “RGD genotype and phenotype resources in modeling of human diseases”
Poster #14: Niran Hadad “Disruption in sleeping behavior in a genetically diverse Alzheimer’s mouse reference panel”
Poster #15: Riyan Cheng “A few small datasets in rats demonstrate the feasibility of low coverage whole genome sequencing”
Poster #16: Saunak Sen “Speeding up eQTL scans in the BXD population”
Poster #17: Spencer Mahaffey “PhenoGen: Building a REST API”
Poster #18: Steve Britt “Genome Wide Association Study of Stochastic Photoreceptor Cell Fate Determination Using the Drosophila Genetic Reference Panel”
Poster #19: Suheeta Roy “Control of longevity by diet versus body weight”
Poster #20: Tristan de Jong “Variation in gene expression, the other dimension in transcriptome analysis”
Poster #21: Won Gyo Suh “Fast linkage methods and approximate significance thresholds for analysis of the Collaborative Cross”
Poster #22: Nikhil Vettikattu “Developmental effects of diethylstilbestrol on mouse bacula”
Poster #23: Xinzhu Zhou “Genome-wide association study in two cohorts from a multi-generational mouse advanced intercross line highlights the difficulty of replication”
3.30 – 5 Poster session with coffee and snacks (rooms 141-145)
5 – 6 Short talks (12 min+3 min QA)
Session chair: Victor Guryev
Celine St. Pierre “Partitioning parent-of-origin- and sequence-dependent effects on allele-specific expression and methylation using a LG/J x SM/J F1 reciprocal cross”
Kathie Sun “Modeling gene-by-environment effects and allelic imbalance in a reciprocal, sparse diallel of Collaborative Cross mice”
Juan Macias-Velasco “Epistatic Networks Associated with Parent-of-Origin Effects on Metabolic Traits”
Amelie Baud “Genetic variants of cage mates contribute to inter-individual phenotypic differences”
Tuesday June 11
Location: Sanford Medical Education and Telemedicine Center, Auditorium (first floor)
8.30 – 9.30 Invited talk: Melissa Gymrek “Dissecting the contribution of tandem repeat variation to complex traits”
9.30 – 10.30 Short talks (12 min+3 min QA)
Session chair: Celine St. Pierre
Danielle Gillard “Susceptibility to Cisplatin Induced Hearing Loss in Mice within the Hybrid Mouse Diversity Panel”
Ely Cheikh Boussaty “Fhod3, The Formin Protein Is Expressed In Mouse Inner Ear And Its Overexpression Leads To Progressive High Frequency Hearing Loss”
Aaron Deal “High Fat diet negatively impacts both metabolic and emotional health in an outbred rat model”
Olivier George “The Addiction Biobanks: two repositories from genetically characterized outbred rats with compulsive-like escalation of cocaine or oxycodone intake”
10.30 – 11 Coffee break
11 – 12.30 Short talks (12 min+3 min QA)
Session chair: Amanda Barkley-Levenson
Cory Parks “Genetic variation in initial sensitivity and rapid desensitization to THC”
Boris L. Tabakoff “The need for a holistic approach to system genetics of drug related behaviour”
Sarah Schoenrock “Initial drug sensitivity and sensitization in the Collaborative Cross and Diversity Outbred mouse populations and their inbred founder strains”
Robert Hitzemann Cancelled
Michael Saul “Reference traits bridge drug use phenotypes and drug-na√Øve brain gene expression in Diversity Outbred mice”
Jonathan Pollock “Funding Opportunities for Genetics at the National Institute on Drug Abuse”
12.30 – 2 Lunch break (no lunch provided). Mentoring will be offered to those interested over lunch. Details TBD.
2 – 3.30 Short talks (12 min+3 min QA)
Session chair: Steven Munger
David Aylor “Gene x environment interactions cause sperm decline and male infertility in mice”
Yanwei Cai “Identifying gene-by-treatment interaction on phenotype, expression and pathway scale using a Collaborative Cross approach”
William Valdar “Determinants of QTL mapping power in the realized Collaborative Cross”
Anna Tyler “Genetic interactions affect lung function in patients with systemic sclerosis”
Matt Mahoney “Sparse genomic prediction models using combined analysis of pleiotropy and epistasis”
Daniel Carlin “Network Assisted Genomic Association: A fast and flexible framework for network assisted genomic association”
3.30 – 4 Coffee break
4 – 5.30 Short talks (12 min+3 min QA)
Session chair: Elissa Chesler
Richard Mott “Pretty Good Genetic Privacy via Orthogonal Transformation”
Greg Keele “Integrative QTL analysis of gene expression and chromatin accessibility identifies multi-tissue patterns of genetic regulation”
Elissa Chesler “Convergence of complex trait regulation across species”
Nikki Russell “The genetic architecture underlying inter-individual variation in the ER stress transcriptional response”
Selcan Aydin “Genetic dissection of the embryonic stem cell proteome”
6 – 9 Conference dinner at Kellogg Park (La Jolla shores)
Use Bus #30 or Uber/Lyft, or ~45 min mostly scenic walk
Workshop: Using GeneNetwork data for your own and good
Sunday June 9, 6-8pm (not overlapping with any other conference session).
Please email [email protected] and [email protected] by Friday May 24 to register for the workshop.
Organizers: Pjotr Prins, Saunak Sen, Karl Broman, Zachary Sloan, Christian Fischer, and Rob W. Williams
GeneNetwork is a rich data and analysis tool for genetically diverse mouse and rat populations. In this tutorial we will introduce the web-based graphical user interface (GUI) of GeneNetwork that allows the user to use powerful built-in tools such as trait correlations, R/qtl, GEMMA, Bayesian Network Analysis, Correlation Trait Loci analysis.
We will demonstrate the new REST (REpresentational State Transfer) that allows R, Python and Julia users the ability to download data in GeneNetwork, and generate customized analyses. We will also show how to fetch data from Wikidata (such as gene names and aliases) to enrich the data analysis. We will use Jupyter Notebooks to present and tweak GeneNetwork functionality live. The notebooks also allow the user to create reproducible and citeable analysis workflows that can be published as mini-publications and research objects.
All important GeneNetwork contributors will be present for the tutorial. Participants are encouraged to interact with the presenters and each other.
See https://trello.com/b/c1u2bjOz/ctc-tutorial for Kanban board with additional information.
Pizza will be provided.
Venue:
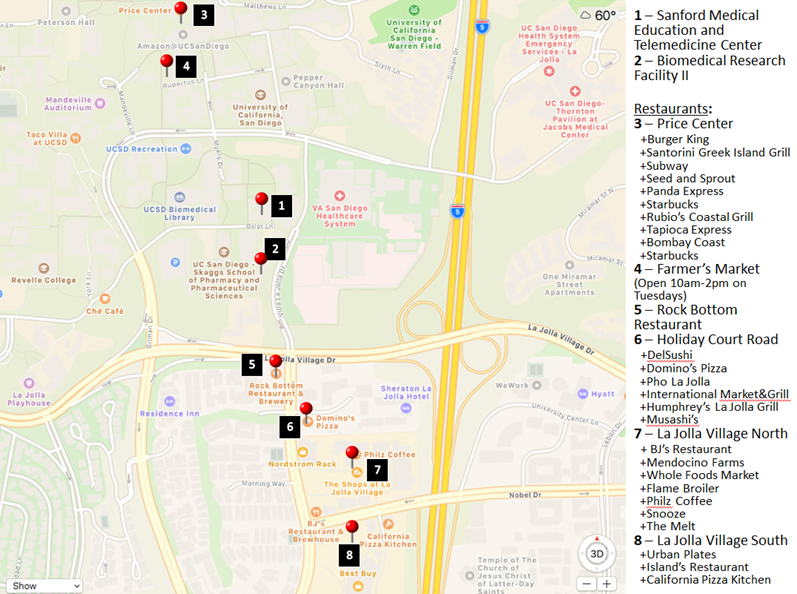
UCSD campus, in Medical Education and Telemedicine building, which is located at the end of Osler lane (see map).
Lactation rooms are available at the venue. Please contact [email protected] for information
Address: the whole campus has the same address (9500 Gilman Dr, La Jolla CA) which will not lead you to the Medical Education and Telemedicine building. You have to search maps for “Medical Education and Telemedicine building, UCSD” to find the place. Make sure you give correct direction to an Uber/Lyft driver.
Meals:
We will provide coffee and snacks, but meals will be the responsibility of attendees. The map below shows several options within walking distance and we will also suggest nearby restaurants that work with services like grubhub, Doordash, UberEats, Amazon Restaurants, Eat24, Postmates, etc.
Hotels:
The hotels nearby include La Jolla Shores Hotel, Estancia La Jolla Hotel & Spa, Sheraton La Jolla Hotel, Hyatt Regency La Jolla At Aventine. You can also use AirB&B to find suitable housing.
Transportation:
From San Diego International Airport (SAN) it is approximately 30 min Uber/Lyft. The whole campus has the same address (9500 Gilman Dr, La Jolla CA) which will not lead you to the Medical Education and Telemedicine building. You have to search maps for “Medical Education and Telemedicine building, UCSD” to find the place. Make sure you give correct directions to your driver.
Public transportation from SAN is possible: use bus 923 or 992, then transfer to express bus 150, it will take approximately 1 hour.
If you would like to drive to the meeting, please be aware that parking on campus is allowed in Visitor spaces, $3/hr. It is best to park elsewhere and then use some transportation option to arrive on campus.
Links for additional files:
For questions, please contact Abraham A. Palmer ([email protected]) and Amelie Baud ([email protected]).
Updated June 5, 2019