We share data in these ways: Internet Rat Server | C-GORD | SRA archive | UC San Diego Library Digital Collections | GitHub | Biobanks | GeneNetwork2

1. The Internet Rat Server (IRS) is a cloud-hosted portal providing the Center’s investigators with easy, password-protected access to the Center’s HS rats database, including an archive of reports and data tools.
(Note: Shiny app was phased out in early 2024)
The Center maintains this database containing all phenotypical data collected by the HS rats research projects and ancillary collaborators, as well as RNAseq and genotype data.
Data requests should be addressed to the Center PI, Abraham Palmer or the Scientific Coordinator, Oksana Polesskaya.

2. C-GORD: The Center for GWAS in Outbred Rats Database (C-GORD) is a publicly-accessible portal to the data generated by the NIDA Center for Genetic Studies of Drug Abuse in Outbred Rats, designed to comply with FAIR (Findable, Accessible, Interoperable, Reusable) principles.
(Note: C-GORD is currently in revision. More information available here.)

3. Sequencing data for low-coverage sequences of HS rats are deposited to SRA, BioProject PRJNA1022514

4. Genotype data from: NIDA Center for GWAS in Outbred Rats from the UC San Diego Library Digital Collections.

5. PalmerLab Github contains releases of genotyping bioinformatics pipelines and other related projects. The content is updated as the lab develops new pipelines.
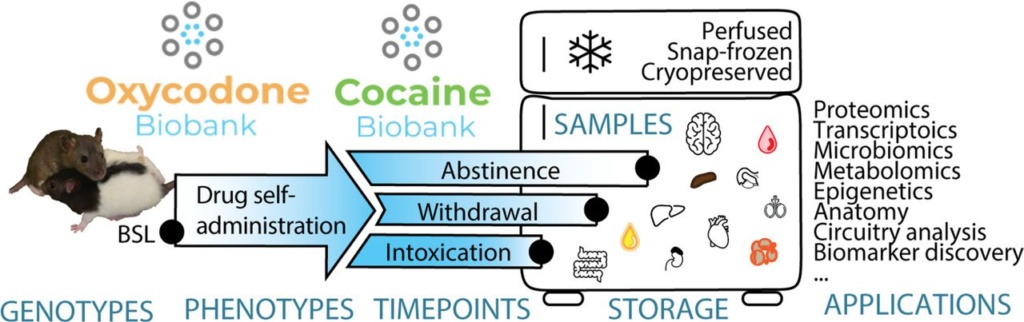
6. Biobanks: We are working with UC San Diego researchers Dr. Olivier George and Dr. Giordano de Guglielmo to provide genotyping for the HS rats that underwent cocaine, oxycodone, or alcohol exposure protocols. The tissues from these animals were collected and preserved in tissue banks and are available for researchers around the world to use in addiction studies: cocaine biobank, oxycodone biobank, alcohol biobank.
The Cocaine and Oxycodone Biobanks, Two Repositories from Genetically Diverse and Behaviorally Characterized Rats for the Study of Addiction.
eNeuro 19 April 2021; 8 (3) ENEURO.0033-21.2021.

7. We are working with the GeneNetwork 2 team at the University of Tennessee Health Science Center to make our data available through this resource. GeneNetwork2 provides access to linked data sets and tools to study complex networks of genes, molecules, and higher-order gene function and phenotypes.
Researchers interested in obtaining additional data from this Center should contact Abraham Palmer.
Updated on:
